Jiang lab
Designing Biologics:
High throughput synthetic biology meets AI
Cell is the fundamental unit of life and the ability to program cells and macromolecules within them holds the key to unlock precision therapeutics for many diseases. The Jiang lab combines synthetic biology and machine learning to develop a molecular technology toolbox for mammalian cell engineering. We take inspirations from natural biological systems and build tools to measure parameters of interest in high throughput format. These datasets fuel the development of deep learning models to help us unravel biological complexity and engineer therapeutics.

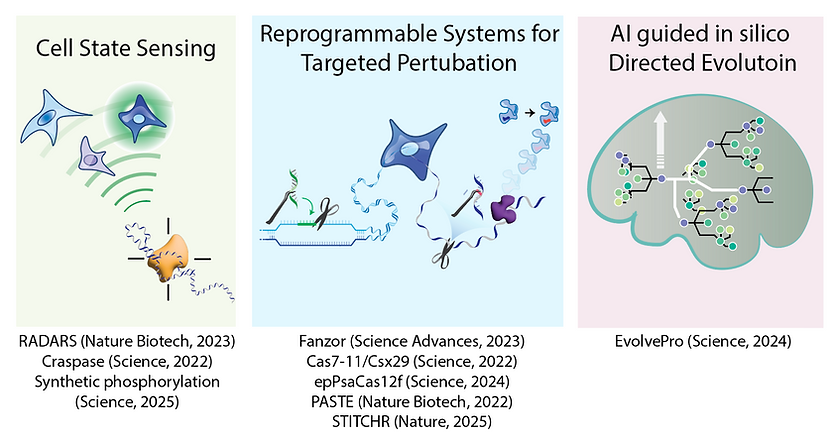
Cell State Sensor

Multiplexed mRNA sensing enables us to sense and manipulate cell state, but technologies so far has been limited by their sensitivity and modularity. Tools like RADARS and Craspase enable programmable and sensitive mRNA detection, but they still lack clear design rules to be broadly applicable. We are collecting high throughput functional data on RNA sensors to develop generative AI models to help us design robust RNA sensors for a variety of applications including lineage tracing to understand cancer evolution, cell therapy for solid tumor and autoimmune disorders, and cell state targeted mRNA therapeutics.
AI for Biology
Current foundation models like ESM and AlphaFold have transformed our understanding of protein. However, to train foundation models that are capable of complex macromolecule design like engineering enzymes, mRNA therapeutics and eventually cells, we need many more high-quality sequences to function data in the right context to train these models. We will bridge the gap by developing high throughput assays to meausre parameters of interest and use them to train predictive models that helps us better design drugs, enzymes, cells and eventually system-level phenotype.

Deep learning guided protein therapeutic design

Protein based therapeutics are among the most important drug modality in modern medicine. The rise of monoclonal antibody like Keytruda and peptide drug like GLP-1 RA enables us to tackle previously intractable disease. However, these type of drugs in general suffer from clinical immunogenicity and lack of design rule during engineering. We are pioneering novel molecular technologies and deep learning model to tackle these hard problems, which will accelerate drug design.
RNA biology and genAI for mRNA therapeutics
Post-transcriptional regulation plays an important role in determining cell state and identity. However, the network of RNA-binding proteins and translational machinery with different mRNA is poorly understood and thus has not been well leveraged for mRNA therapeutic design. We are combining high throughput sequencing approaches with deep learning genAI models to understand the complexity of this regulation and design cell state targeted RNA therapeutics.

Synthetic Immunity

Current cell therapy are typically manufactured ex vivo with viral vector and thus they are hard to engineer, slow and expansive. RNA based cell state sensor technologies allow us to sense and manipulate cell state to overcome challenges like T cell exhaustion in solid tumor and cytokine storms. Combining proteo-lipid nanoparticle with mRNA circuits will allow us to generate synthetic immunity to treat a wide range of diseases
Understanding Cancer Evolution
Cell state plasticity is a key feature of cancer and the ability to dynamically switch cell state and cell fate drives therapeutic resistance and metastasis. End point measurements like scRNA-seq and mass-spec have unveiled heterogeneity of cancer but they do not capture the dynamics of cancer evolution that are critical in disease intervention and therapeutic design. Recent rise in lineage tracing technologies has revealed the phylogenies of cancer evolution and metastasis, but we still do not fully understand the precise cell state trajectories that they take. In addition, we do not have molecular technology to label and manipulate cell state of interest during this process. In light of this, we are deploying cell state sensor technologies in the context of lung cancer to better understand the development of these cells and perform targeted perturbation to inform better therapeutic design.
